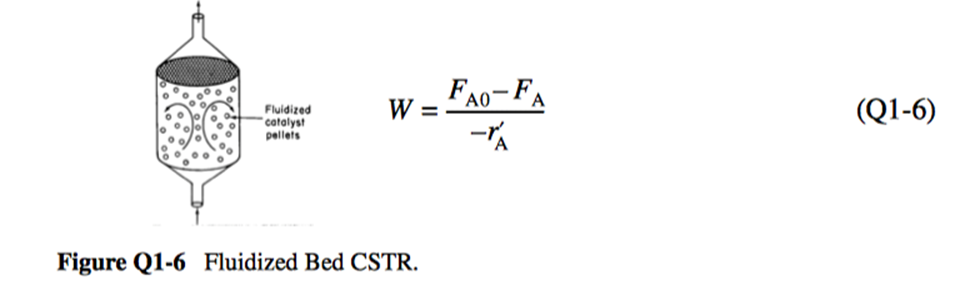Workshop 01: mole balances
Lecture notes for chemical reaction engineering
Solutions to these problems are uploaded at Workshop 1 solutions
Try following problems from Fogler 5e(Fogler 2016).
Q 1-5, Q 1-6, P 1-4, P 1-5, P 1-8.
We will go through some of these problems in the workshop.
Q 1-5: What assumptions were made in the derivation of the design equation for:
- The batch reactor (BR)?
- The CSTR?
- The plug-flow reactor (PFR)?
- The packed-bed reactor (PBR)?
- State in words the meanings of and .
Q 1-6: Use the mole balance to derive an equation analogous to Equation (1-7) ( for a fluidized CSTR containing catalyst particles (Figure 1) in terms of the catalyst weight, , and other appropriate terms.
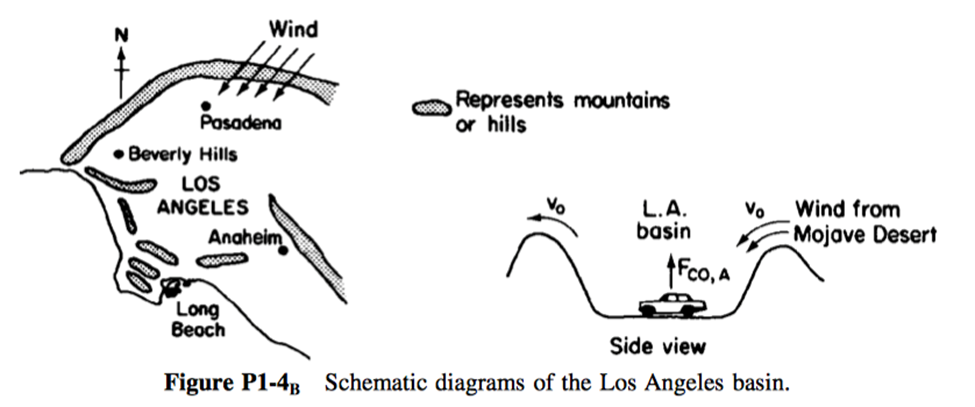
P 1-4: Schematic diagrams of the Los Angeles basin are shown in Figure 2. The basin floor covers approximately 700 square miles ( ) and is almost completely surrounded by mountain ranges. If one assumes an inversion height in the basin of 2,000 ft, the corresponding volume of air in the basin is ( ). We shall use this system volume to model the accumulation and depletion of air pollutants. As a very rough first approximation, we shall treat the Los Angeles basin as a well-mixed container (analogous to a CSTR) in which there are no spatial variations in pollutant concentrations.
We shall perform an unsteady-state mole balance (Equation (1–4) ()) on as it is depleted from the basin area by a Santa Ana wind. Santa Ana winds are high-velocity winds that originate in the Mojave Desert just to the northeast of Los Angeles. Load the Smog in Los Angeles Basin Web Module. Use the data in the module to work parts 1–12 (a) through
- given in the module. Load the Living Example Polymath code and explore the problem. For part (i), vary the parameters , a, and b, and write a paragraph describing what you find.
There is heavier traffic in the L.A. basin in the mornings and in the evenings as workers go to and from work in downtown L.A. Consequently, the flow of CO into the L.A. basin might be better represented by the sine function over a 24-hour period.
P 1-5: The reaction is to be carried out isothermally in a continuous-flow reactor. The entering volumetric flow rate is . (Note: . For a constant volumetric flow rate , then . Also,
Calculate both the CSTR and PFR reactor volumes necessary to consume 99% of A (i.e., ) when the entering molar flow rate is , assuming the reaction rate is
- with
- with
- with
- Repeat (a), (b), and/or (c) to calculate the time necessary to consume 99.9% of species A in a constant-volume batch reactor with .
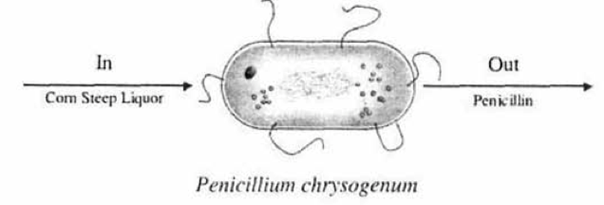
We are going to consider the cell as a reactor. The nutrient corn steep liquor enters the cell of the microorganism Penicillium chrysogenum (Figure 3) and is decomposed to form such products as amino acids. RNA. and DNA. Write an unsteady mass balance on
- the corn steep liquor.
- RNA, and
- penicillin. Assume the cell is well mixed and that RNA remains inside the cell.
References
Citation
@online{utikar2024,
author = {Utikar, Ranjeet},
title = {Workshop 01: Mole Balances},
date = {2024-02-24},
url = {https://cre.smilelab.dev/content/workshops/01-mole-balances/},
langid = {en}
}