Collection and analysis of rate data
Chemical Reaction Engineering
Rate data analysis
- Isothermal reactor design algorithm
Isothermal reactor design algorithm
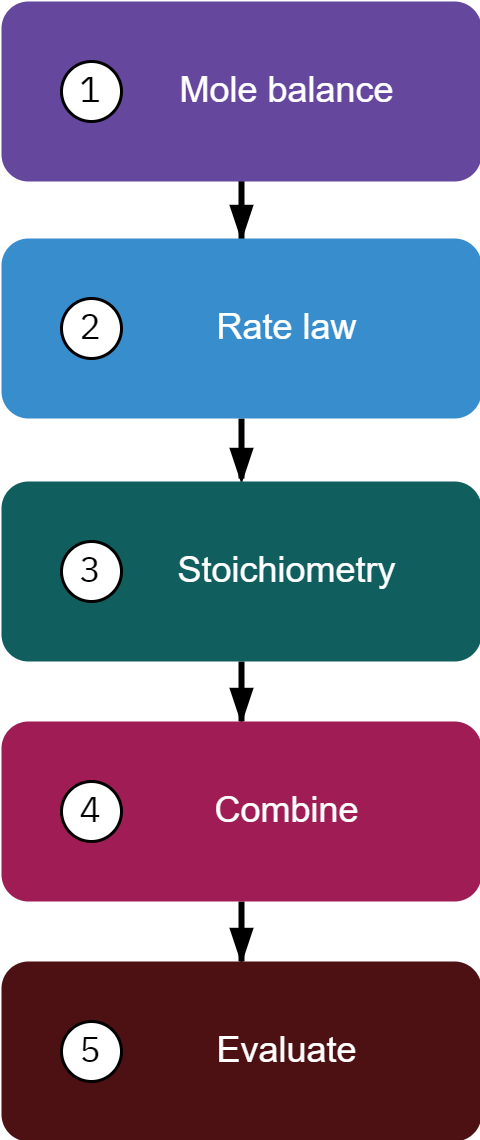
Mole balance
FA0−FA=∫VrAdV=dtdNA
Rate law
If −rA is given as f(X) → directly solve the design equations
Stoichiometry
If −rA=g(C) → use stoichiometry to write −rA=f(X)
Combine
Gather all equations to obtain a system of equations that must be solved.
Evaluate
The system of equation scan be solved analytically, graphically, numerically, or using software
Isothermal reactor design: molar flow rates
In many instances it is easier to work with molar flow rates/ no. of moles than conversion
- Reactors with external mass transfer (such as membrane reactors)
- Multiple reactions in gas phase
We must write a mole balance for each and every species as opposed to just one species
Usually this leads to a system of (simultaneous or) ordinary differential equations
Solve the combined set of equations using ODE solver (such as
scipy.integrate.solve_ivp)
Isothermal reactions in PBR: molar flow rates
Second order reaction aA+bBcC+dD

Mole balance: Write balance for each species i=1 to N
dWdFi=ri′
Rate law
−rA′=kCAαCBβ;−arA′=−brB′=crC′=drD′
Stoichiometry Ci=(1+ϵX)CA0(Θi+νiX)(P0P)(TT0)
Pressure: dWdP=−2pα(T0T)FT0FT
Total molar flow rate: FT=∑i=1NFi
Combine:
Collate all equations from steps 1 to 3 to yield a system of equations
Evaluate:
Use ODE solver to solve the system of equations obtained in step 4.
Reactor design problem

In practice, collection and analysis of rate data is the most time consuming task in reactor design

Data collection is done in the lab, where we can simplify mole balance, stoichiometry, and fluid dynamic considerations
Rate law
Algebraic equation that relates −rA to species concentration
−rA=kA(T)f(CA,CB,...)
Power law model
- Most common general forms of the rate law
−rA=kACAαCBβ
Order of reaction: the powers to which the concentrations are raised
- The reaction is α order with respect to A and β order with respect to B.
- Overall order of reaction n=α+β.
kA: Specific reaction rate
- Depends on the temperature
Need to determine k, α, and β
Collection and analysis of rate data
Goal
- Determine reaction order, α, and specific reaction rate constant, k, in the rate law
- Want ideal conditions → well-mixed (data is easiest to interpret) → Select a simple reactor
Constant-volume batch reactor
- For homogenous reactions
- Concentration vs. time measurements
- Measurement during the unsteady-state operation
Differential reactor
- For solid-fluid reactions
- Measurement during steady state operation
- Product concentration is usually monitored for different feed conditions
Algorithm for rate data analysis
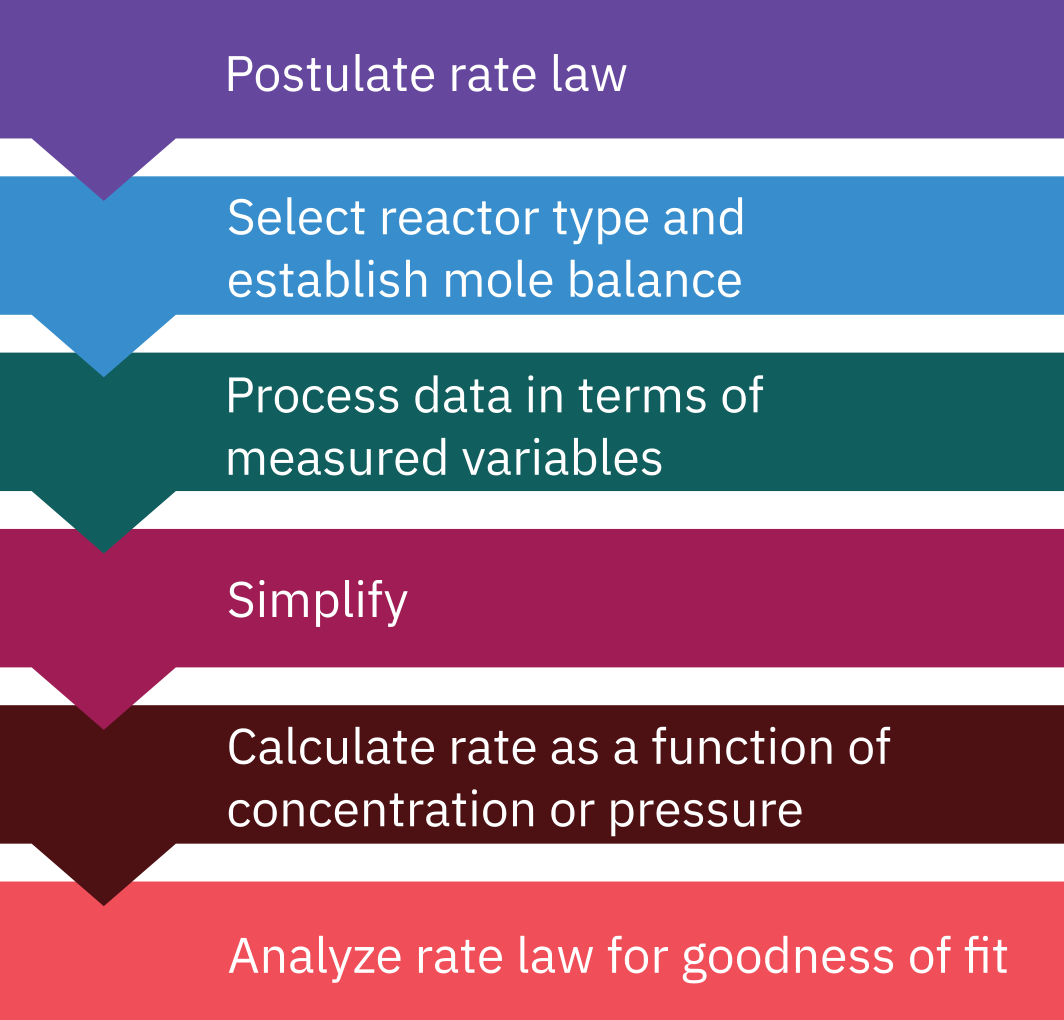
Determination of rate law for homogenous reactions
Most often batch reactors are used
Type of reactor chosen will not affect rate of reaction
Batch reactor
- Simple operations, low cost
- Ease of sampling, easy clean up, limited waste
- Uniform concentration can be obtained
Mole balance: constant volume −dtdCA=−rA
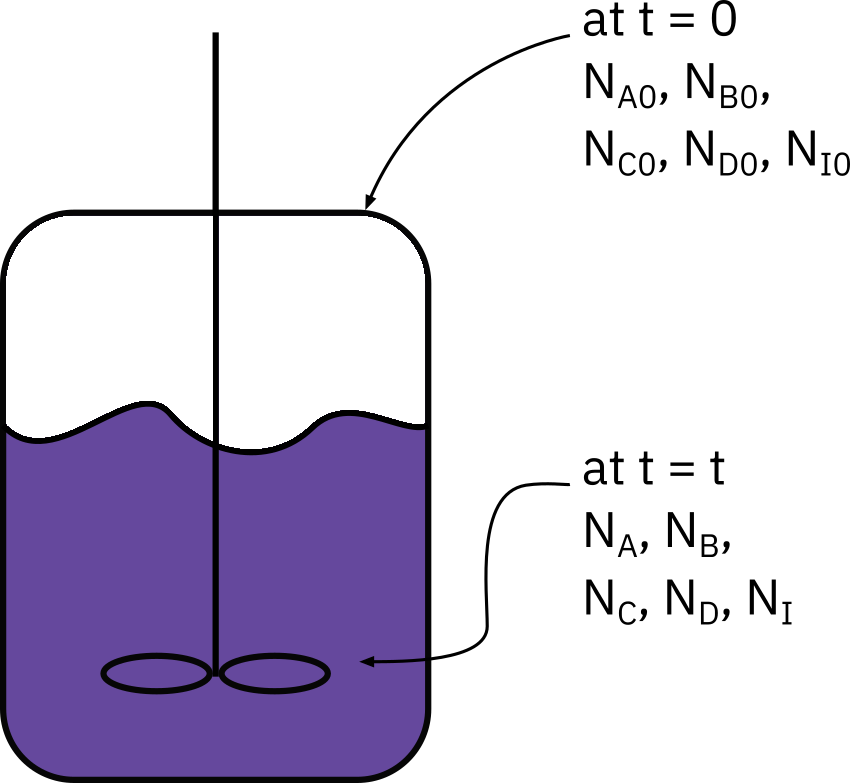
- Typical measurements
- Concentration Pressure
- Temperature: Many times batch reactions are carried out isothermally.
- Development of heat during reaction: Reaction calorimetry
Method of excess
Given concentration vs. time profile in a batch experiment, determine the reaction order and rate constant.
A+BC+D
- Rate law: −rA=kCAαCBβ
- Need to determine: k, α, and β

Determining reaction orders: α, and β
Common simplification: One of the reactants is in excess
Two separarate experiments
Excess B ⇒ CB≫CA, CB can be assumed constant. ⇒ determine α.
Excess A ⇒ CA≫CB, CA can be assumed constant. ⇒ determine β.
Determining k: Measuring rate at known concentrations of A, and B.
Analysis methods
- Differential analysis
- Integral analysis
- Nonlinear regression
- Method of half life
- Initial rates method
- Differential reactor
Differential analysis
Irreversible reaction
The rate is essentially a function of the concentration of only one reactant.
Aproducts;−rA=kCAα
Isothermal, constant volume batch reactor
Mole balance: constant volume −dtdCA=−rA;−rA=kCAα
Taking natural logarithm
ln(dt−dCA)=lnk+αlnCA
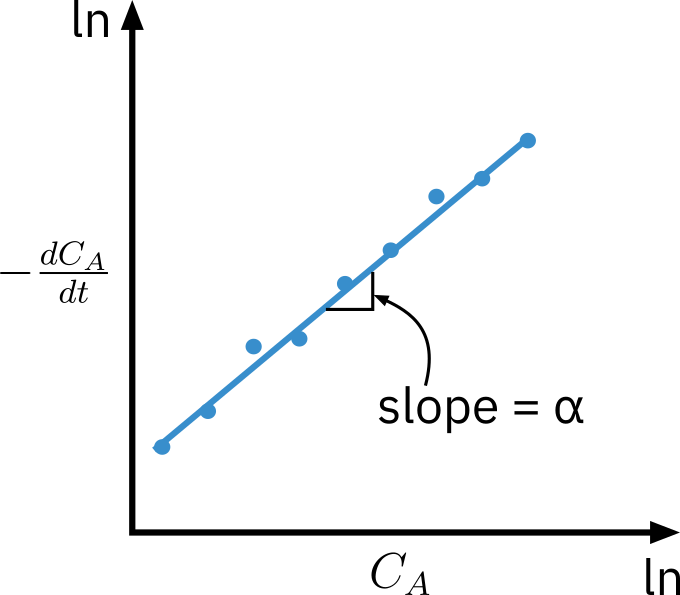
Slope of plot of ln[−dCA/dt] vs.lnCA is the reaction order
Specific reaction rate can be determined using a specific point p: k=(CA)pα(−dCA/dt)p
Evaluating dt−dCA
Graphical diffrentiation
- Very old method
- Equal area graphical diffrentiation
- Disparities in the data are easily seen
Numerical diffrentiation
- Finite difference
- Independent variable (time) is equally spaced
Diffrentiation of a polynomial fit to the data
Fit a polynomial to CA vs. t data
e.g. CA=f(t)=a0+a1t+a2t2+a3t3+a4t4
Analytical derivative
dtdCA=f′(t)=a1+a2t+a3t2+a4t3
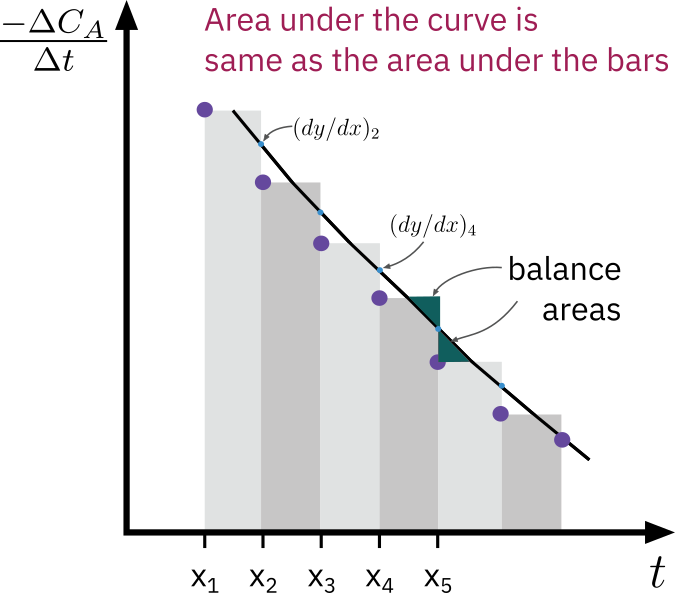
- Determine reaction order and specific rate from plot of ln[−dCA/dt] vs.lnCA
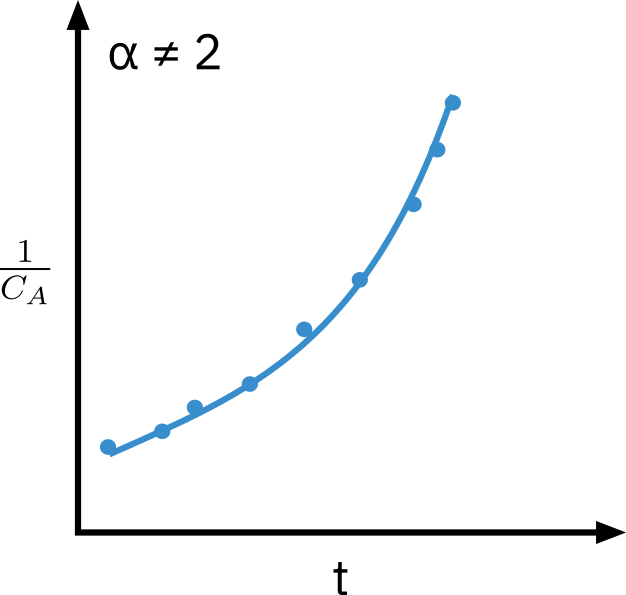
Integral analysis
Quickest method to determine if the order is 0, 1, or 2.
⇒ Used when reaction order can be guessed or is known.
Guess reaction order
Integate the differential equation to obtain concentration as a function of time
If the guessed order is correct, appropriate plot (determined from integration) should be linear.
Mole balance: −dCA/dt=−rA
Zero order reaction
−rA=k
CA=CA0−kt
First order reaction
−rA=kCA
lnCACA0=kt
Second order reaction
−rA=kCA2
CA1=CA01+kt
Need to know appropriate function of concentration corresponding to a rate law that is linear with time.
Integral analysis
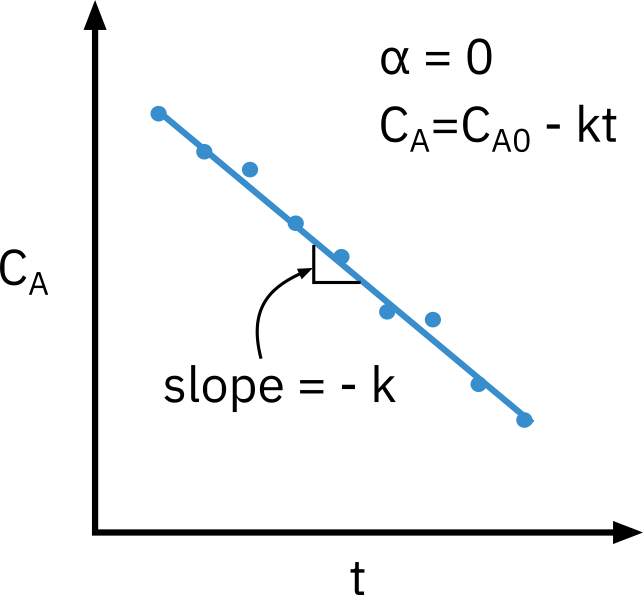
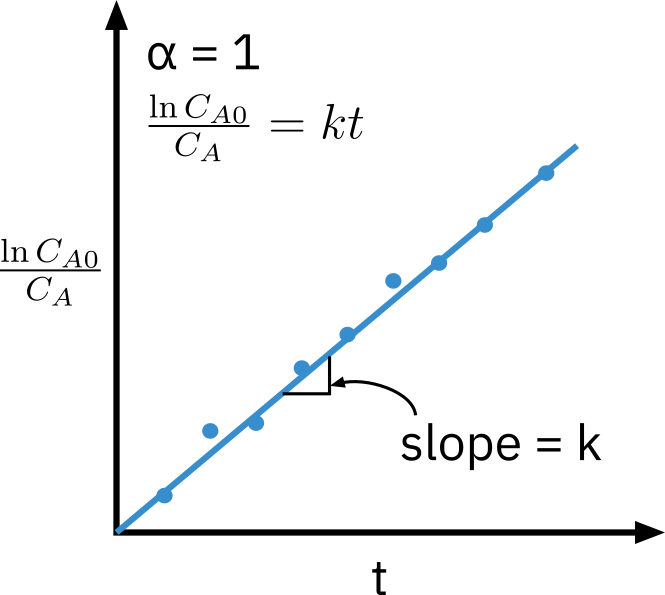
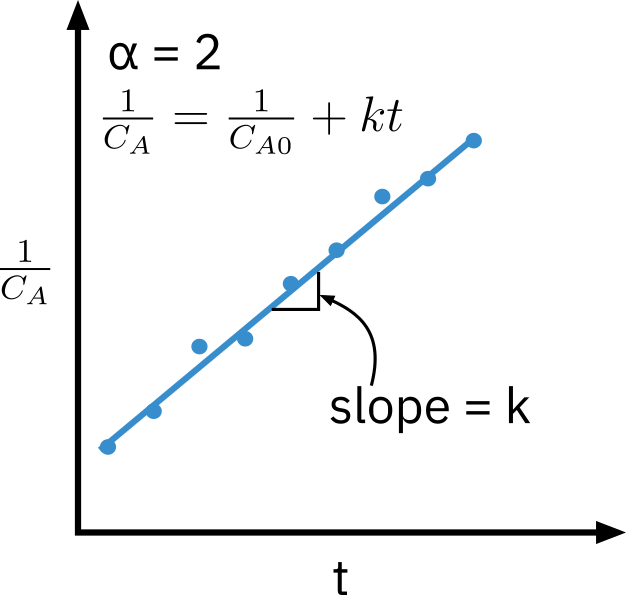
Nonlinear regression
Search for parameter values that minimize the sum of squares of the difference between the measured values and calculated values for all data points.
Best estimate of parameter values
Discriminate between different rate law models
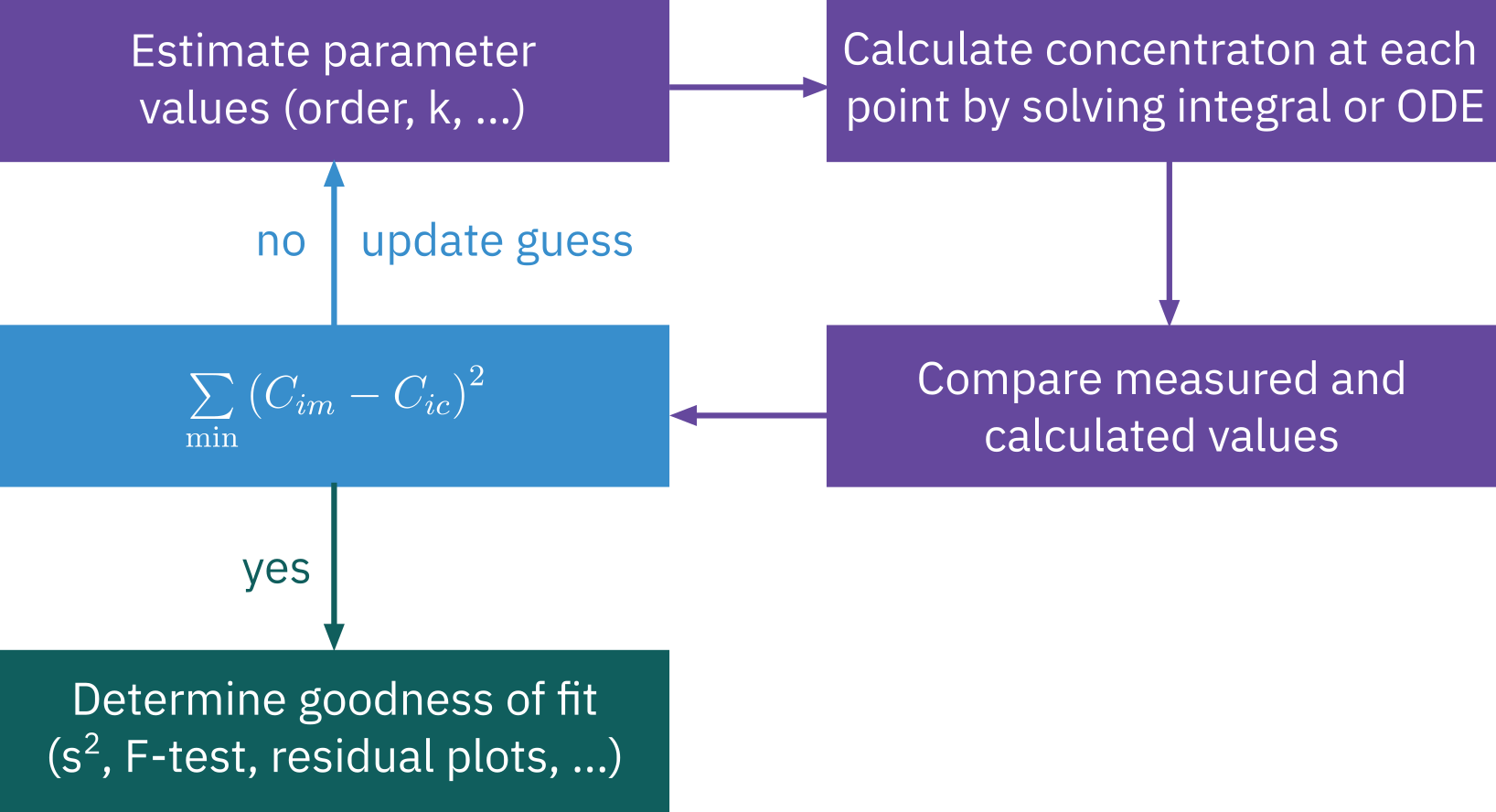
Godness of fit
Root Mean Squared Error (RMSE) and Mean Absolute Error (MAE)
Lower values indicate a better fit. RMSE is sensitive to outliers, while MAE provides a more robust error metric.
P-value of the F-test in ANOVA (Analysis of Variance)
A p-value smaller than the significance level (commonly 0.05) indicates that there is a statistically significant relationship.
Residual Plots
Ideally, the residuals should be randomly scattered around 0 across the range of fitted values. Patterns in the residual plot can indicate problems with the model
Method of half life
Half life (t1/2): Time taken for the concentration of reactant to fall to half of its initial value.
Determine half life as a function of initial concentration
Requires several experiments
Reaction: Aproducts
Rate law: −rA=kCAα
Mole balance: −dtdCA=kCAα
- Integration: CA=CA0 at t=0
t=kCA0α−1(α−1)1[(CACA0)α−1−1]
- Integration: CA=21CA0 at t=t1/2
t1/2=kCA0α−1(α−1)1[2α−1−1]
- In general:
t1/n=k(α−1)nα−1−1[CA0α−11]
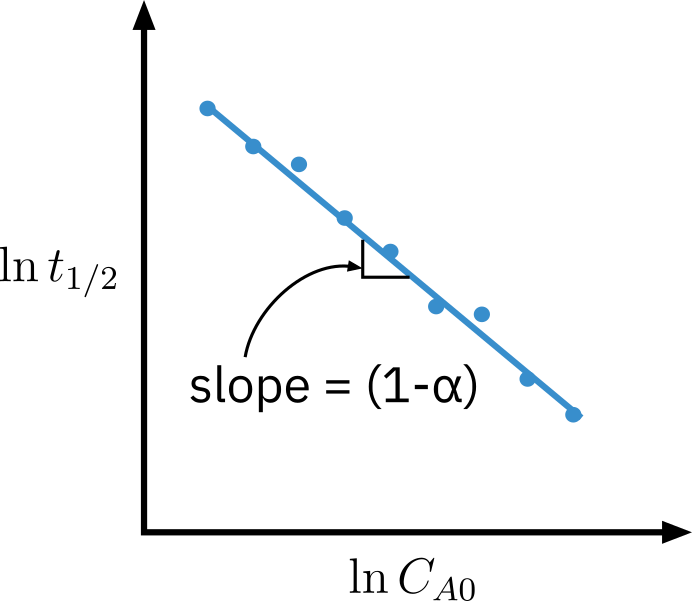
Method of half life
t1/2=kCA0α−1(α−1)1[2α−1−1]
Taking log
lnt1/2=ln(α−1)k2α−1−1+(1−α)lnCA0
Multiple experiments are performed varying initial concentration and t1/2 is recorded.
The plot of lnt1/2 vs. lnCA0 is linear with a slope of (1−α)
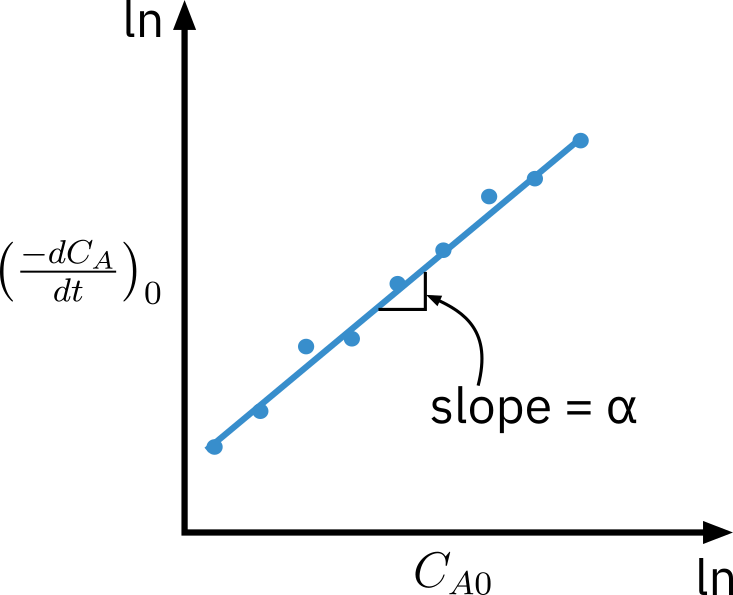
Method of initial rates
Perform a series of experiments at different initial concentrations CA0
Determine initial rate of reaction −rA0
Determine rate law parameters by relating −rA0 to CA0
Reaction: Aproducts
Rate law: −rA=kCAα
Mole balance: −dtdCA=kCAα
Initial rate: −rA0=(dt−dCA)0=kCA0α
- Taking log
ln(dt−dCA)0=lnk+αlnCA0
Rate data from differential reactors
For heterogeneous reactions mostly packed bed reactors (PBRs) are used.
Differential reactor: The conversion of the reactants in the bed is extremely small, as is the change in reactant concentration through the bed
Reactant concentration through the reactor is essentially constant (i.e. the reactor is considered to be gradient-less)
Can treat the mole balance like a CSTR
Rate of reaction determined for a specified number of pre-determined initial or entering reactant concentrations
Determine rate of reaction as a function concentration or partial pressure
Operate isothermally
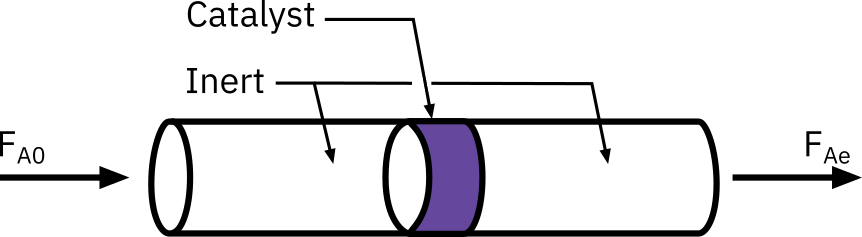
Differential reactor
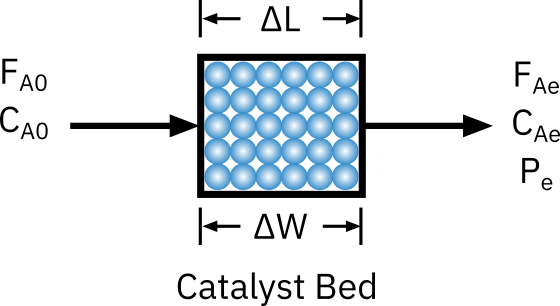
Assumptions
- No concentration and temperature gradient (gradientless reactor)
- High volumetric flow rate
- Small catalyst particles (No mass transfer limitation)
- Very low conversion
- Low/ negligible heat release (isothermal)
- No bypassing/ channeling (uniform flow across catalyst layer)
Mole balance:
in - out + generation = accumulation
FA0−FAe+rA′ΔW=0
−rA′=ΔWFA0−FAe=ΔWυ0CA0−υCAe
- For constant flow rate υ=υ0
−rA′=ΔWυ0CA0X=ΔWυ0CP
→ For small conversion −rA′ can be expressed as a function of CA0
This component is an instance of the CodeMirror interactive text editor. The editor has been configured so that the Tab key controls the indentation of code. To move focus away from the editor, press the Escape key, and then press the Tab key directly after it. Escape and then Shift-Tab can also be used to move focus backwards.

Chemical Reaction Engineering